During drug development, it’s critical to monitor cellular responses to promising compounds, which includes keeping tabs on the activities of various gene promoters. Researchers from the RIKEN Center for Life Science Technologies report a new method to accomplish that, using the advanced gene expression analysis method CAGE followed by single-molecule sequencing.
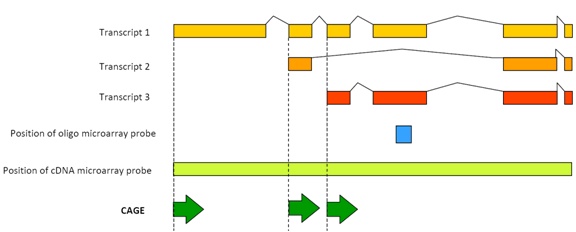
Click to enlarge. Original image and CAGE details can be found at the RIKEN website: http://www.osc.riken.jp/english/activity/cage/basic/
This new technique, published in CPT: Pharmacometrics & Systems Pharmacology, September 2013, features a more precise analysis of cellular responses to drugs, at the level of individual promoters, than the microarray-based methods that are commonly employed. Dr. Harukazu Suzuki and his team developed their new technique by combining Cap Analysis of Gene Expression (CAGE), which was also developed at RIKEN, with 3rd generation, single-molecule sequencing.
CAGE sequences the 5’-end of mRNAs to comprehensively map human transcription start sites and their promoters and quantify the set of mRNAs, or transcriptome, in a cell, and reveal information about the level of expression of specific genes. The RIKEN team used CAGE, combined with a single-molecule sequencer, to monitor the effect of three drugs on human breast cancer cells. The study identified a distinct set of promoters that were altered by low doses of the drugs at a level of sensitivity that would be very difficult using other profiling techniques.
These results illustrate the potential utility of highly quantitative promoter activity profiling for drug research and development.
To learn more about the Gene Expression Profiling market, access our Gene Expression Profiling Dashboard here. Or contact us at info@perceptaassociates.com to discuss a custom project.


Leave a Reply