As has been shown again and again, seemingly homogeneous cell populations often consist of more than one cell type, which can confound research results. Single-cell research offers a bottom-up approach to circumventing such unwanted cellular heterogeneity. As a testament to its potential promise, the NIH has committed ~$90 million over 5 years to single cell analysis research. At Percepta, we were curious to know which “omic” method of single-cell analysis is the most compelling to researchers. We posed this question to a portion of our scientific panel who are currently engaged in cellular localization studies:
Q: “Which, if any, of the following single-cell methods (Single Cell Genomics, Single Cell Proteomics, Single Cell Metabolomics, or Single Cell Transcriptomics) do you believe to be most powerful and/or you are most excited about?”
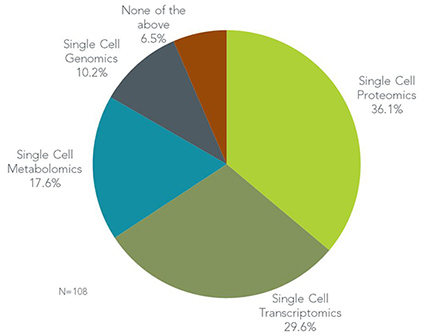 As shown in the Figure, a majority (36.1%) of cell localization scientist indicate single-cell proteomics is the most powerful/exciting. Researchers expressed the sentiment that “protein expression gets closer to predicting functional cellular outcomes” over other omics. In addition, they told us protein expression is “more correlative to mechanism of action” and that they are the “best” to target and validate. Single cell proteomics is also suggested as a powerful tool for protein network/pathway analysis, particularly when using a “heavily multiplexed analysis”.
As shown in the Figure, a majority (36.1%) of cell localization scientist indicate single-cell proteomics is the most powerful/exciting. Researchers expressed the sentiment that “protein expression gets closer to predicting functional cellular outcomes” over other omics. In addition, they told us protein expression is “more correlative to mechanism of action” and that they are the “best” to target and validate. Single cell proteomics is also suggested as a powerful tool for protein network/pathway analysis, particularly when using a “heavily multiplexed analysis”.
Single-cell transcriptomics runs as a close second to single-cell proteomics and was cited by 29.6% of respondents for “real transcriptome profiling” of single cells rather than cell populations. This approach is considered useful for understanding tumor heterogeneity, transcriptional modification or alternative transcript isoforms. Further, it is a viable option when samples are very limited or precious.
Though single-cell metabolomics was cited by only 17.6% of cell localization researchers, some protagonists indicated that it is a “fascinating” and “growing science”, and may be most likely to produce new data to show “the whole picture” of what’s really going on in the cell.


Leave a Reply